drord: Doubly robust estimators of ordinal treatment effects
David Benkeser
Emory University
Department of Biostatistics and Bioinformatics
1518 Clifton Road, NE
Mailstop: 002-3AA
Atlanta, Georgia, 30322, U.S.A.
benkeser@emory.edu
Source: Emory University
Department of Biostatistics and Bioinformatics
1518 Clifton Road, NE
Mailstop: 002-3AA
Atlanta, Georgia, 30322, U.S.A.
benkeser@emory.edu
vignettes/using_drord.Rmd
using_drord.RmdAbstract
Many treatments are evaluated using randomized trials or observational studies examining effects or associations with ordinal outcomes. In these situations, researchers must carefully select a statistical estimand that reflects an interpretable and clinically relevant treatment effect. Several such estimands have been proposed in the literature. In the context of randomized trials, simple nonparametric estimators are available. These estimators can be improved through adjustment for covariates that are prognostic of the study outcome. The drord package implements covariate-adjusted estimators of several popular estimands for evaluating treatment effects on ordinal outcomes. The estimators use working models for the treatment probability as a function of covariates (i.e., the propensity score) and the outcome distribution as a function of covariates. The latter is based on a proportional odds model. Estimates of these two quantities are combined to provide estimates of treatment effects. The treatment effect estimates are doubly robust, in that they are consistent if one of the two working models contains the truth. The effect estimates are nonparametric efficient if both working models contain the truth. This vignette provides a brief introduction to the drord package, demonstrating several estimators that are available therein.
Introduction
Ordinal outcomes are commonly encountered in practice. For example, randomized trials evaluating treatments for COVID-19, the disease caused by the SARS-CoV-2 virus, often consider ordinal outcomes such as: death, intubation, or no adverse event. In these settings it is important to pre-specify an estimand that quantifies the effect of the treatment on the outcome distribution in a way that is interpretable and closely related to patient care. Several such estimands are commonly employed including:
- Difference in (weighted) means: The outcome levels are assigned a numeric value and possibly additionally assigned a weight reflecting that some outcomes may be considerably worse than others. The effect is defined as the difference in weighted average outcomes between two treatments.
- Log odds ratio: The comparison describes the average over all levels of the log-odds ratio comparing two treatments of the cumulative probability for each level of the outcome. See (Díaz, Colantuoni, and Rosenblum 2016) for further discussion.
- Mann-Whitney: The probability that a randomly-selected individual receiving a given treatment will have a more favorable outcome than a randomly selected individual receiving an alternative treatment (with ties assigned weight 1/2). See (Vermeulen, Thas, and Vansteelandt 2015) for further discussion.
Simple nonparametric estimates of these quantities are readily available. For example, in the context of a randomized trial, the difference in means parameter can be consistently estimated by a difference in sample means. Similarly straightforward estimates are available for the other quantities. These simple estimators can be improved by considering adjustment for baseline covariates that are prognostic of the study outcome. Adjusting for covariates yields estimates that are more precise than unadjusted estimates. Thus, the power to detect treatment effects may be improved by utilizing these estimators. The drord package provides covariate-adjusted estimates of the three quantities above, along with several methods that are useful for analyzing studies with ordinal outcomes.
Estimators
To solidify ideas, we briefly introduce notation. Let \(X\) be a vector of baseline covariates that could include continuous, binary, or categorical components, \(A\) be a binary treatment vector (assuming values 1, e.g., for active treatment and 0, e.g., for a control treatment or standard of care), and \(Y\) be an ordinal-valued outcome encoded to assume numeric values \(j = 0, 1, ..., K\). We assume that the data consist of \(n\) independent copies of the random variable \(O = (X,A,Y)\).
Our estimators are constructed by first estimating key nuisance parameters describing different aspects of the distribution of \(O\). These nuisance parameters are not directly of interest, but are used as an intermediate step in the estimation procedure. We describe these nuisance quantities mathematically and return to these ideas when we demonstrate how to control their estimation within the drord function.
Nuisance parameters
The first nuisance parameter is \(P(A | X)\), the conditional probability of the treatment variable given baseline covariates. This quantity is known exactly in a randomized trial and may not depend at all on \(X\) (e.g., if randomization is not stratified by covariates). We use \(\hat{\pi}(a | x)\) to denote an estimate of \(P(A = a | X = x)\) for \(a = 0, 1\) and a given covariate values \(x\). This estimate could be set exactly equal to the known randomization probability, the sample proportion that receive treatment \(a\), or a logistic regression model to adjust for chance imbalances in \(X\) across the treatment arms.
We also require estimates of \(\{ P(Y \le j | A = a, X), j = 0, 1, ..., J \}\) for \(a = 0, 1\). We refer to these as treatment-specific conditional distribution functions (CDFs) of the outcome. Our estimators are built on a working proportional odds regression model: for \(j = 0, ..., K-1\), \[ \mbox{logit}\{P(Y \le j | A = a, X = x)\} = m_{\alpha_a, \beta_a}(j,x) = \alpha_a(j) + \beta_a^\top x \ . \] Importantly, the validity (i.e., consistency, asymptotic normality) of our estimators does not rely on this working model being correctly specified. For \(a = 0, 1\) our estimates of \(\alpha_a, \beta_a\) are chosen by minimizing \[ -\sum_{j=0}^{K-1} \sum_{i=1}^n \frac{I\{A_i = a\}}{\hat{\pi}(A_i | X_i)} \log\left[m_{\alpha,\beta}(j,X_i)^{I(Y_i\le j)}\{1-m_{\alpha,\beta}(j,X_i)\}^{I(Y_i> j)}\right] \ . \label{eq:empiricalRisk} \] This estimation can be carried out using standard software packages (discussed in detail below) for fitting proportional odds models by including observation-level weights equal for the \(i\)-th observation equal to \(I\{A_i = a\}/\hat{\pi}(A_i | X_i)\). Note that this procedure is repeated in each treatment arm separately.
Finally, we require an estimate for each \(x\) of \(F_X(x) = P(X \le x)\), the distribution of baseline covariates. Our estimators use the empirical distribution of this quantity.
Parameters of interest
We can create a covariate-adjusted estimate of the CDF \(\psi_a(j) = P(Y\le j|A=a)\) by marginalizing the conditional CDF estimates with respect to the distribution of baseline covariates. By the law of total probability \(\psi_a(j) = E\{P(Y \le j | A = a, X)\} = \int P(Y \le j | A = a, X) dF_X(x)\). A plug-in estimate of this quantity based on our working model estimates is \[ \hat{\psi}_a(j) = \frac{1}{n}\sum_{i=1}^n m_{\hat{\alpha}_a,\hat{\beta}_a}(j,X_i). \label{eq:CDF} \]
This estimate of \(\psi_a(j)\) is doubly robust in that it is consistent if at least one of \(\hat{\pi}\) and \(\mbox{logit}^{-1}(m_{\hat{\alpha}_a, \hat{\beta}_a})\) are consistent forthe true propensity score or true conditional CDF, respectively. The key to establishing this is to note that by including weights in the proportional odds model and including \(j\)-specific intercept terms \[ \frac{1}{n}\sum_{i=1}^n \frac{I\{A_i=a\}}{\hat{\pi}(A_i|X_i)}\left\{I(Y_i\le j) - m_{\hat{\alpha}_a,\hat{\beta}_a}(j,X_i)\right\} = 0. \] is satisfied for each \(j = 0, ..., K\).
The estimates \(\hat{\psi}_a(j)\) of \(\psi_a(j)\) can now be mapped into estimates of the effects of interest. For simplicity of notation, we introduce notation for the implied treatment-specific probability mass function (PMF). In particular, we define \(\hat{\theta}_a(0) = \hat{\psi}_a(0)\) and for \(j = 1, \dots, K\) define \(\hat{\theta}_a(j) = \hat{\psi}_a(j) - \hat{\psi}_a(j-1)\). Estimates of the treatment effect parameters can be computed as follows. The estimate for the difference in weighted means is \[ \sum_{j=0}^K \ j \ w_j \{\hat{\theta}_1(j) - \hat{\theta}_0(j)\} \ , \] where \(w_j\) is the weight assigned to outcome level \(j\). The estimate of the average log-odds ratio is \[ \frac{1}{K} \sum_{j=0}^{K-1} \left[\mbox{log}\left\{\frac{\hat{\psi}_1(j)}{1 - \hat{\psi}_1(j)} \right\} - \mbox{log} \left\{\frac{\hat{\psi}_0(j)}{1 - \hat{\psi}_0(j)} \right\} \right] \ . \] The estimate of the Mann-Whitney estimand is \[ \sum_{j=0}^{K} \left\{\hat{\psi}_0(j-1) + \frac{1}{2} \hat{\theta}_0(j) \right\} \hat{\theta}_1(j) \ . \]
Standard error estimates can be derived using influence functions and the delta method. Alternatively, nonparametric bootstrap can be used. In the next section, we discuss the implementation of these for building confidence intervals.
Using the drord function
COVID-19 data
The main function in this package is the eponymous drord function. We illustrate its usage using the covid19 data set included with the package. This is a simulated data set mimicking a randomized COVID-19 treatment trial, where we will illustrate how such a trial could be analyzed while adjusting for patient age. The control arm data were simulated to match the outcomes of a CDC preliminary description of 2,449 cases reported to the CDC from February 12 to March 16, 2020. The data were simulated such that the treatment decreased the probability of intubation, but has no impact on the probability of death. The data can be loaded and viewed as follows.
## out age_grp treat
## 1 3 3 1
## 2 3 6 0
## 3 2 7 0The data include three variables:
-
out: the study outcome with 1 representing death, 2 intubation, 3 no adverse outcome; -
age_grp: age category with 1 the youngest and 7 the oldest represent age groups (0-19, 20-44, 45-54, 55-64, 65-74, 75-84, \(\ge\) 85); -
treat: hypothetical treatment, here 1 represents an the effective, active treatment and 0 a standard of care condition.
Basic calls and plots
We begin with a simple call to drord to familiarize ourselves with its output
(fit1 <- drord(out = covid19$out, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE]))## $mann_whitney
## est wald_cil wald_ciu
## 0.5800689 0.5337929 0.6263449
##
## $log_odds
## est wald_cil wald_ciu
## treat1 -1.2922742 -1.5719616 -1.01258678
## treat0 -0.9163279 -1.1533670 -0.67928881
## diff -0.3759463 -0.7254384 -0.02645421
##
## $weighted_mean
## est wald_cil wald_ciu
## treat1 2.5554260 2.46316500 2.6476871
## treat0 2.3721986 2.28768451 2.4567127
## diff 0.1832275 0.06389974 0.3025552The main function inputs are out, treat, and covar. Note that covar must be input as a data.frame even though in this example it contains only a single covariate. We see that the function returns estimates of each of the three parameters described above. For the log-odds and weighted mean parameters, output is broken down into treatment-specific estimates, as well as the difference. Thus the row labeled treat1 in $weighted_mean corresponds to inference about \(\sum_{j=0}^K \ j \ w_j \theta_1(j)\), the weighted mean in the treat=1 group. Similarly, the row label treat0 in $log_odds corresponds to inference about \[
\frac{1}{K} \sum_{j=0}^{K-1} \mbox{log}\left\{\frac{\psi_1(j)}{1 - \psi_1(j)} \right\} \ .
\]
In addition to the treatment effect parameters, one can obtain inference about \(\psi_a\) and \(\theta_a\). This information can be viewed in the $cdf and $pdf output, respectively, or can be plotted using ggplot2 via the plot.drord method.
# plot of CDF
cdf_plot <- plot(fit1, dist = "cdf",
treat_labels = c("Treatment", "Control"),
out_labels = c("Death", "Death or intubation"))
cdf_plot$plot + ggsci::scale_fill_nejm()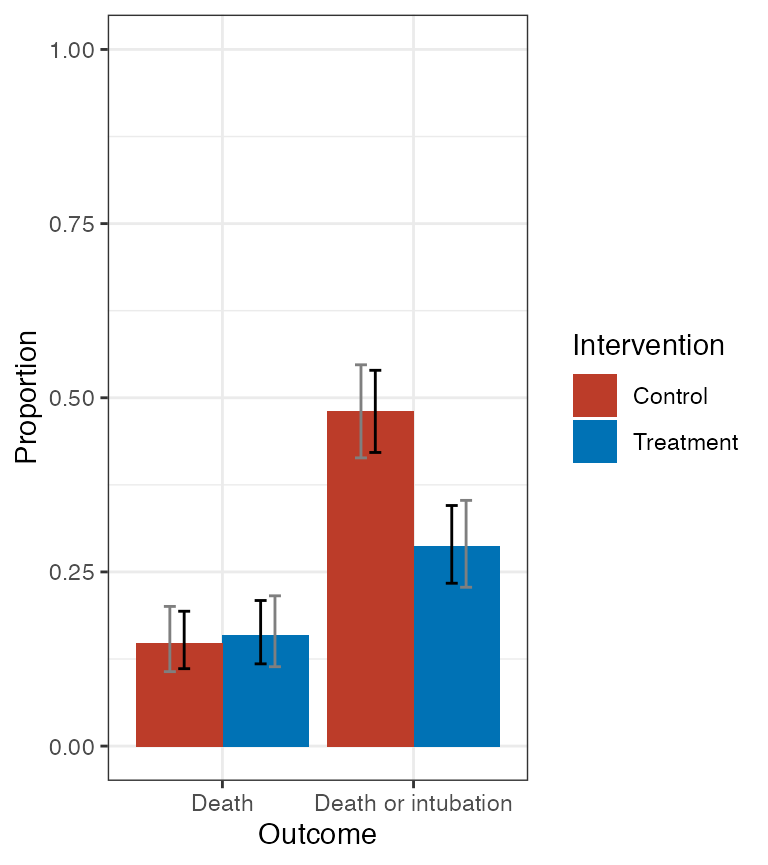
Plot of treatment-specific CDF. Black bars are pointwise 95% confidence intervals; gray bars are 95% simultaneous confidence bands.
Note that custom labels for treatments and outcome levels are allowed. For the CDF, the the outcome labels should correspond to labels for outcome levels 0, 1, …, \(K-1\) (as the CDF evaluated at \(K\) is always equal to 1).
A similar plot for the PMF can be obtained.
# plot of PMF
pmf_plot <- plot(fit1, dist = "pmf",
treat_labels = c("Treatment", "Control"),
out_labels = c("Death", "Intubation", "None"))
pmf_plot$plot + ggsci::scale_fill_nejm() 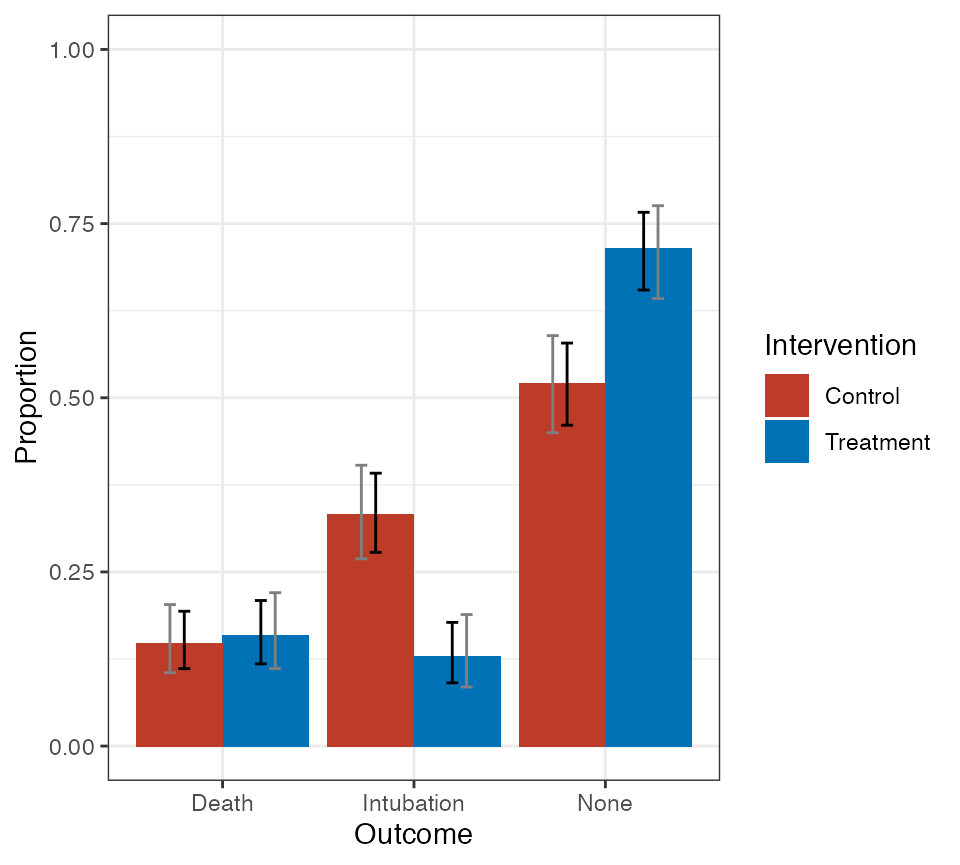
Plot of treatment-specific PMF. Black bars are pointwise 95% confidence intervals; gray bars are 95% simultaneous confidence bands.
Text can be added to the bars as follow.
pmf_plot$plot +
ggsci::scale_fill_nejm() +
ggplot2::geom_text(ggplot2::aes(y = 0,
label = sprintf("%1.2f", Proportion)),
position = ggplot2::position_dodge(0.9),
color = "white",
vjust = "bottom")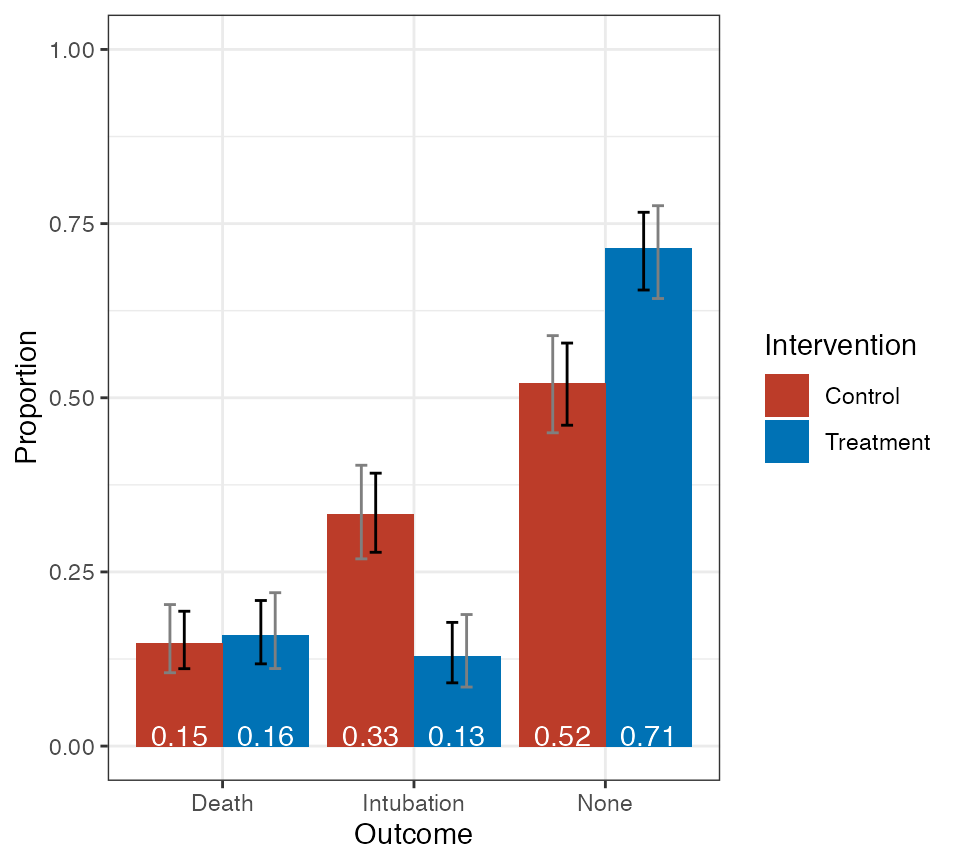
Plot of treatment-specific PMF with text added.
Working model controls
In the simple call, we used default working models. The drord documentation indicates that the working model fits can be controlled via the out_form and treat_form models. These options include the right-hand-side of a regression formula for estimation of their respective nuisance parameters. The default for the outcome model (out_form = ".")is a main-effects pooled logistic regression model for proportional odds. The default for the treatment model is an intercept-only logistic regression model (treat_form = 1), i.e., the sample proportion of individuals in each treatment arm. Here, we illustrate another call to drord where we treat age_grp as a factor rather than numeric in the outcome model and include adjustment for age_grp in the propensity model.
(fit2 <- drord(out = covid19$out, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE],
out_form = "factor(age_grp)",
treat_form = "factor(age_grp)"))## $mann_whitney
## est wald_cil wald_ciu
## 0.5795097 0.5337040 0.6253155
##
## $log_odds
## est wald_cil wald_ciu
## treat1 -1.3037590 -1.5715576 -1.03596052
## treat0 -0.8903684 -1.1365746 -0.64416223
## diff -0.4133906 -0.7594801 -0.06730116
##
## $weighted_mean
## est wald_cil wald_ciu
## treat1 2.5563878 2.46842001 2.6443556
## treat0 2.3665194 2.27816560 2.4548732
## diff 0.1898684 0.07113711 0.3085997By default models are returned in the drord object; this behavior can be suppressed via the return_models option. The fitted working models can be accessed via $out_mod and $treat_mod, which we can use to confirm that the models were fit as expected in fit1 and fit2. Note that out_mod is a list of length two, which contains a fitted proportional odds model for each treatment level.
# age_grp treated as numeric in fit1
fit1$out_mod$treat1## $glm_fit
##
## Call: stats::glm(formula = stats::update(form, stats::formula(paste0(new_out_name,
## "~.-1+", out_name))), family = binomial(), data = data_pooled,
## weights = wgts)
##
## Coefficients:
## age_grp out1 out2
## 0.696 -5.570 -4.693
##
## Degrees of Freedom: 482 Total (i.e. Null); 479 Residual
## Null Deviance: 1386
## Residual Deviance: 910.5 AIC: 883.7
##
## $data
## out age_grp
## 1 3 3
## 5 1 6
## 9 3 4
## 10 3 3
## 12 3 3
## 14 2 6
## 15 3 7
## 17 3 4
## 18 3 3
## 20 3 7
## 24 3 6
## 27 1 6
## 28 3 3
## 32 3 4
## 34 3 4
## 35 3 2
## 37 3 2
## 39 2 5
## 41 2 7
## 44 3 5
## 45 3 3
## 46 3 3
## 47 2 3
## 50 3 4
## 52 3 4
## 55 3 5
## 58 3 3
## 59 3 7
## 60 3 3
## 61 1 6
## 62 3 7
## 63 3 5
## 66 3 7
## 67 3 3
## 69 3 7
## 75 3 7
## 79 3 6
## 81 3 5
## 82 2 5
## 83 3 2
## 90 3 3
## 95 3 4
## 97 3 3
## 98 1 7
## 99 3 7
## 108 3 6
## 109 3 6
## 111 2 5
## 114 3 7
## 116 3 7
## 117 3 3
## 119 3 7
## 122 3 3
## 124 2 7
## 127 3 4
## 129 1 4
## 131 3 7
## 132 2 6
## 135 3 2
## 137 3 5
## 140 2 4
## 142 3 7
## 144 3 5
## 145 3 6
## 150 3 6
## 153 3 7
## 162 3 5
## 163 3 4
## 165 3 6
## 166 3 6
## 169 2 5
## 175 1 7
## 176 3 3
## 181 3 1
## 183 1 7
## 184 3 6
## 185 3 7
## 187 1 7
## 188 3 4
## 189 3 7
## 190 3 5
## 192 3 1
## 194 2 6
## 195 3 7
## 198 3 3
## 201 3 6
## 203 2 6
## 206 1 7
## 207 2 7
## 208 1 7
## 209 3 5
## 210 3 3
## 213 1 7
## 215 3 4
## 216 3 6
## 218 3 4
## 219 3 5
## 222 3 7
## 223 1 7
## 224 1 7
## 225 3 6
## 226 3 4
## 227 3 7
## 230 3 6
## 233 3 3
## 234 1 6
## 235 3 6
## 236 3 3
## 238 3 5
## 241 3 5
## 243 3 3
## 244 3 7
## 247 1 7
## 249 3 6
## 250 2 6
## 251 2 6
## 254 1 7
## 257 1 7
## 260 1 6
## 261 3 7
## 263 3 6
## 264 3 2
## 267 2 5
## 268 3 3
## 270 1 7
## 272 3 7
## 273 3 7
## 274 1 7
## 275 1 7
## 276 3 4
## 278 1 7
## 279 2 5
## 280 1 6
## 281 3 3
## 285 3 3
## 294 3 4
## 295 3 7
## 296 1 7
## 298 3 5
## 299 2 6
## 300 3 6
## 302 3 3
## 306 3 4
## 307 3 4
## 310 2 7
## 311 1 7
## 312 2 6
## 313 3 4
## 316 3 7
## 318 2 7
## 319 3 4
## 322 3 5
## 323 3 4
## 324 3 2
## 332 3 7
## 337 3 5
## 338 3 7
## 341 3 5
## 342 3 6
## 343 3 7
## 344 3 7
## 346 3 7
## 351 3 1
## 352 3 6
## 354 3 5
## 355 3 2
## 356 2 6
## 358 3 3
## 362 2 7
## 365 3 4
## 370 1 7
## 372 3 2
## 373 3 6
## 374 3 5
## 376 2 6
## 377 1 5
## 378 3 4
## 380 3 5
## 382 3 1
## 383 3 4
## 384 3 7
## 385 1 7
## 387 3 7
## 389 3 2
## 392 3 1
## 393 3 3
## 397 3 6
## 399 3 5
## 400 3 3
## 401 3 3
## 402 3 5
## 403 3 4
## 404 3 3
## 406 3 6
## 413 3 4
## 414 3 2
## 416 3 4
## 417 1 7
## 421 2 3
## 422 1 6
## 423 3 5
## 424 3 6
## 426 3 6
## 427 1 7
## 430 2 7
## 432 3 2
## 438 1 7
## 439 3 3
## 444 3 3
## 445 1 3
## 447 3 6
## 449 3 6
## 452 3 7
## 453 3 7
## 454 2 4
## 457 3 2
## 461 1 6
## 462 3 3
## 463 2 6
## 464 1 7
## 465 3 3
## 468 3 3
## 472 1 7
## 473 3 6
## 474 2 4
## 477 2 4
## 479 3 7
## 480 2 3
## 481 3 4
## 482 3 7
## 484 3 6
## 485 1 6
## 486 1 7
## 490 3 7
## 491 3 7
## 493 1 7
## 495 3 3
## 496 2 5
## 497 1 7
## 499 3 7
## 500 1 7
##
## $levs
## [1] "1" "2" "3"
##
## $out_name
## [1] "out"
##
## attr(,"class")
## [1] "POplugin"
# age_grp treated as factor in fit2
fit2$out_mod$treat1## $glm_fit
##
## Call: stats::glm(formula = stats::update(form, stats::formula(paste0(new_out_name,
## "~.-1+", out_name))), family = binomial(), data = data_pooled,
## weights = wgts)
##
## Coefficients:
## factor(age_grp)1 factor(age_grp)2 factor(age_grp)3 factor(age_grp)4
## -18.3822 -18.5538 -3.2663 -2.8477
## factor(age_grp)5 factor(age_grp)6 factor(age_grp)7 out2
## -2.3014 -1.3696 -0.7694 0.9703
##
## Degrees of Freedom: 482 Total (i.e. Null); 474 Residual
## Null Deviance: 1386
## Residual Deviance: 873.7 AIC: 935.5
##
## $data
## out age_grp
## 1 3 3
## 5 1 6
## 9 3 4
## 10 3 3
## 12 3 3
## 14 2 6
## 15 3 7
## 17 3 4
## 18 3 3
## 20 3 7
## 24 3 6
## 27 1 6
## 28 3 3
## 32 3 4
## 34 3 4
## 35 3 2
## 37 3 2
## 39 2 5
## 41 2 7
## 44 3 5
## 45 3 3
## 46 3 3
## 47 2 3
## 50 3 4
## 52 3 4
## 55 3 5
## 58 3 3
## 59 3 7
## 60 3 3
## 61 1 6
## 62 3 7
## 63 3 5
## 66 3 7
## 67 3 3
## 69 3 7
## 75 3 7
## 79 3 6
## 81 3 5
## 82 2 5
## 83 3 2
## 90 3 3
## 95 3 4
## 97 3 3
## 98 1 7
## 99 3 7
## 108 3 6
## 109 3 6
## 111 2 5
## 114 3 7
## 116 3 7
## 117 3 3
## 119 3 7
## 122 3 3
## 124 2 7
## 127 3 4
## 129 1 4
## 131 3 7
## 132 2 6
## 135 3 2
## 137 3 5
## 140 2 4
## 142 3 7
## 144 3 5
## 145 3 6
## 150 3 6
## 153 3 7
## 162 3 5
## 163 3 4
## 165 3 6
## 166 3 6
## 169 2 5
## 175 1 7
## 176 3 3
## 181 3 1
## 183 1 7
## 184 3 6
## 185 3 7
## 187 1 7
## 188 3 4
## 189 3 7
## 190 3 5
## 192 3 1
## 194 2 6
## 195 3 7
## 198 3 3
## 201 3 6
## 203 2 6
## 206 1 7
## 207 2 7
## 208 1 7
## 209 3 5
## 210 3 3
## 213 1 7
## 215 3 4
## 216 3 6
## 218 3 4
## 219 3 5
## 222 3 7
## 223 1 7
## 224 1 7
## 225 3 6
## 226 3 4
## 227 3 7
## 230 3 6
## 233 3 3
## 234 1 6
## 235 3 6
## 236 3 3
## 238 3 5
## 241 3 5
## 243 3 3
## 244 3 7
## 247 1 7
## 249 3 6
## 250 2 6
## 251 2 6
## 254 1 7
## 257 1 7
## 260 1 6
## 261 3 7
## 263 3 6
## 264 3 2
## 267 2 5
## 268 3 3
## 270 1 7
## 272 3 7
## 273 3 7
## 274 1 7
## 275 1 7
## 276 3 4
## 278 1 7
## 279 2 5
## 280 1 6
## 281 3 3
## 285 3 3
## 294 3 4
## 295 3 7
## 296 1 7
## 298 3 5
## 299 2 6
## 300 3 6
## 302 3 3
## 306 3 4
## 307 3 4
## 310 2 7
## 311 1 7
## 312 2 6
## 313 3 4
## 316 3 7
## 318 2 7
## 319 3 4
## 322 3 5
## 323 3 4
## 324 3 2
## 332 3 7
## 337 3 5
## 338 3 7
## 341 3 5
## 342 3 6
## 343 3 7
## 344 3 7
## 346 3 7
## 351 3 1
## 352 3 6
## 354 3 5
## 355 3 2
## 356 2 6
## 358 3 3
## 362 2 7
## 365 3 4
## 370 1 7
## 372 3 2
## 373 3 6
## 374 3 5
## 376 2 6
## 377 1 5
## 378 3 4
## 380 3 5
## 382 3 1
## 383 3 4
## 384 3 7
## 385 1 7
## 387 3 7
## 389 3 2
## 392 3 1
## 393 3 3
## 397 3 6
## 399 3 5
## 400 3 3
## 401 3 3
## 402 3 5
## 403 3 4
## 404 3 3
## 406 3 6
## 413 3 4
## 414 3 2
## 416 3 4
## 417 1 7
## 421 2 3
## 422 1 6
## 423 3 5
## 424 3 6
## 426 3 6
## 427 1 7
## 430 2 7
## 432 3 2
## 438 1 7
## 439 3 3
## 444 3 3
## 445 1 3
## 447 3 6
## 449 3 6
## 452 3 7
## 453 3 7
## 454 2 4
## 457 3 2
## 461 1 6
## 462 3 3
## 463 2 6
## 464 1 7
## 465 3 3
## 468 3 3
## 472 1 7
## 473 3 6
## 474 2 4
## 477 2 4
## 479 3 7
## 480 2 3
## 481 3 4
## 482 3 7
## 484 3 6
## 485 1 6
## 486 1 7
## 490 3 7
## 491 3 7
## 493 1 7
## 495 3 3
## 496 2 5
## 497 1 7
## 499 3 7
## 500 1 7
##
## $levs
## [1] "1" "2" "3"
##
## $out_name
## [1] "out"
##
## attr(,"class")
## [1] "POplugin"Other packages are available for fitting the proportional odds models, though for technical reasons, these approaches are not generally recommended. Each of the three proportional odds implementations that are available are based on the same model, but the numerical routines for estimating model parameters are different. Thus, if warnings or errors appear in the ordinal fitting routine, one may consider one of the alternative packages. Options are vglm (from the VGAM package), or polr (from the MASS package). In simulations, we have seen more stable performance from vglm and clm. Here we demonstrate this control.
# use vglm to fit model instead
fit3 <- drord(out = covid19$out, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE],
out_model = "vglm")
# view model output
fit3$out_mod$treat1##
## Call:
## VGAM::vglm(formula = stats::as.formula(paste0("factor(out, ordered = TRUE) ~",
## out_form)), family = VGAM::propodds, data = data.frame(out = out,
## covar)[treat == trt_level, , drop = FALSE], weights = 1/trt_spec_prob_est[treat ==
## trt_level])
##
##
## Coefficients:
## (Intercept):1 (Intercept):2 age_grp
## 5.1677586 4.3064221 -0.6384325
##
## Degrees of Freedom: 482 Total; 479 Residual
## Residual deviance: 731.7883
## Log-likelihood: -365.8941Bootstrap confidence intervals
By default, drord will used closed-form inference. While this is generally faster, we may instead prefer to use bootstrap confidence intervals, which should provide more robust behavior when working models are misspecified. Bootstrap intervals can be requested via the ci = "bca", with option nboot determining the number of bootstrap resamples performed. The bootstrap employed by the package is the bias corrected and accelerated bootstrap (Efron and Tibshirani 1994). In practice, we recommend using many bootstrap samples (e.g., nboot = 1e4), so that inference is not dependent on the random seed that is set; however, here we illustrate using a small number of bootstrap samples for fast compilation of this vignette. To increase speed, we also turn off estimation of confidence intervals for the CDF and PMF by setting est_dist = FALSE.
(fit4 <- drord(out = covid19$out, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE],
ci = "bca", nboot = 20, # use more bootstrap samples in practice!!!
param = "mann_whitney", # only compute mann-whitney estimator
est_dist = FALSE)) # save time by not computing CIs for CDF/PMF## $mann_whitney
## est bca_cil bca_ciu
## 0.5800689 0.5471615 0.6146912Stratified estimators
An alternative approach in settings with low-dimensional covariates is to use fully stratified estimators. That is, we take our estimate of \(P(Y \le j | A = a, X = x)\) to be the empirical proportion of the sample with \(A = a\) and \(X = x\) observed to have \(Y \le j\). This is the nonparametric maximum likelihood estimator. The estimator is nonparametric efficient under no assumptions; however, this estimator can only be computed when \(X\) contains only discrete components. Moreover, it will exhibit unstable finite-sample behavior if only few observations are observed with \(A = a\) and \(X = x\). Thus, our implementation of this estimator is restricted to univariate \(X\). If, in reality, \(X\) contains e.g., 2 binary components, then a univariate covariate could be constructed with one level for each of the cells in the 2x2 table. We leave this sort of pre-processing to the user.
Here we illustrate the stratified estimator in action.
(fit5 <- drord(out = covid19$out, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE],
stratify = TRUE))## $mann_whitney
## est wald_cil wald_ciu
## 0.5795097 0.5335462 0.6254733
##
## $log_odds
## est wald_cil wald_ciu
## treat1 -1.3037591 -1.5875619 -1.01995619
## treat0 -0.8903684 -1.1215380 -0.65919891
## diff -0.4133906 -0.7617605 -0.06502069
##
## $weighted_mean
## est wald_cil wald_ciu
## treat1 2.5563878 2.46409511 2.6486805
## treat0 2.3665194 2.28201897 2.4510198
## diff 0.1898684 0.07065993 0.3090769In general, out_form is ignored if stratify = TRUE; the exception is if out_form = "1", then it is assumed that one wishes to obtain a covariate-unadjusted estimate of the treatment effects.
(fit6 <- drord(out = covid19$out, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE],
stratify = TRUE, out_form = "1",
param = "weighted_mean"))## $weighted_mean
## est wald_cil wald_ciu
## treat1 2.5269710 2.429936910 2.6240050
## treat0 2.3938224 2.306190947 2.4814538
## diff 0.1331486 0.002401185 0.2638959
# compare to sample means
(samp_mean_treat1 <- mean(covid19$out[covid19$treat == 1]))## [1] 2.526971
(samp_mean_treat0 <- mean(covid19$out[covid19$treat == 0]))## [1] 2.393822Missing outcome values
Missing outcome values are permitted in drord. To implement this approach, the function simply applies the methods as described above, but with treat recoded as 0 to indicate that an observation had treat = 0 and had their outcome measured, 1 to indicate an observation that had treat = 1 and had their outcome measured, and -1 to indicate that the outcome is missing. The propensity score estimation thus includes two components, one for the probability that the re-coded treat = 1 given covar, the other for the probability that the re-coded treat = 0 given covar. In the example below, we introduce some missingness into the outcome based on age_grp and call drord.
# missingness probability
miss_out_prob <- plogis(-2 + as.numeric(covid19$age_grp < 5))
miss_out <- rbinom(nrow(covid19), 1, miss_out_prob) == 1
out_with_miss <- covid19$out
out_with_miss[miss_out] <- NA
# correctly model missingness
(fit7 <- drord(out = out_with_miss, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE],
treat_form = "I(age_grp < 5)"))## $mann_whitney
## est wald_cil wald_ciu
## 0.5612776 0.5122419 0.6103134
##
## $log_odds
## est wald_cil wald_ciu
## treat1 -1.1955037 -1.4844206 -0.9065868
## treat0 -1.0008050 -1.2638434 -0.7377667
## diff -0.1946987 -0.5698264 0.1804291
##
## $weighted_mean
## est wald_cil wald_ciu
## treat1 2.5249679 2.423789789 2.626146
## treat0 2.4025430 2.311798726 2.493287
## diff 0.1224249 -0.008130262 0.252980Risk difference for binary outcomes
Though not the focus of the drord package, it is possible to use drord to compute a doubly robust estimate of the risk difference when the outcome is binary. In this case, the package defaults to estimating the outcome distribution using logistic regression (as implemented in the stats::glm function). We demonstrate in the call below.
# collapse categories to make a binary outcome
covid19_binary_out <- as.numeric(covid19$out == 3)
# call drord, only requesting weighted mean with equal
# weights; which in this case equals the risk difference
(fit8 <- drord(out = covid19_binary_out, treat = covid19$treat,
covar = covid19[ , "age_grp", drop = FALSE],
param = "weighted_mean",
est_dist = FALSE)) # must be FALSE to run## $weighted_mean
## est wald_cil wald_ciu
## treat1 0.7108203 0.6549009 0.7667397
## treat0 0.5214215 0.4622688 0.5805742
## diff 0.1893988 0.1107782 0.2680193
# confirm that glm was used for outcome model
class(fit8$out_mod$treat1)## [1] "glm" "lm"Session Info
## R version 4.0.3 (2020-10-10)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS High Sierra 10.13.4
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] drord_1.0.1
##
## loaded via a namespace (and not attached):
## [1] tidyselect_1.1.0 xfun_0.21 bslib_0.2.4.9000
## [4] purrr_0.3.4 lattice_0.20-41 splines_4.0.3
## [7] colorspace_2.0-0 vctrs_0.3.6 generics_0.1.0
## [10] htmltools_0.5.1.9000 stats4_4.0.3 yaml_2.2.1
## [13] utf8_1.1.4 rlang_0.4.10 pkgdown_1.6.1
## [16] jquerylib_0.1.3 pillar_1.5.0 glue_1.4.2
## [19] DBI_1.1.1 lifecycle_1.0.0 stringr_1.4.0
## [22] munsell_0.5.0 gtable_0.3.0 ggsci_2.9
## [25] ragg_1.1.2 VGAM_1.1-5 memoise_2.0.0
## [28] evaluate_0.14 labeling_0.4.2 knitr_1.31
## [31] fastmap_1.1.0 fansi_0.4.2 highr_0.8
## [34] scales_1.1.1 cachem_1.0.4 desc_1.2.0
## [37] jsonlite_1.7.2 farver_2.1.0 systemfonts_1.0.1
## [40] fs_1.5.0 textshaping_0.3.4 ggplot2_3.3.3
## [43] digest_0.6.27 stringi_1.5.3 dplyr_1.0.4
## [46] ordinal_2019.12-10 numDeriv_2016.8-1.1 grid_4.0.3
## [49] rprojroot_2.0.2 tools_4.0.3 magrittr_2.0.1
## [52] sass_0.3.1.9000 tibble_3.1.0 ucminf_1.1-4
## [55] crayon_1.4.1 pkgconfig_2.0.3 MASS_7.3-53.1
## [58] ellipsis_0.3.1 Matrix_1.3-2 assertthat_0.2.1
## [61] rmarkdown_2.7 R6_2.5.0 compiler_4.0.3